Home > Structure > Department of Gene Expression > Developmental Epigenetics
Developmental Epigenetics
Our lab aims at understanding the biological importance of the 3D organization of the genome within the nucleus. We study developmental epigenetics and DNA replication.

Michał Gdula, Phd DSc
Principal Investigator
Research
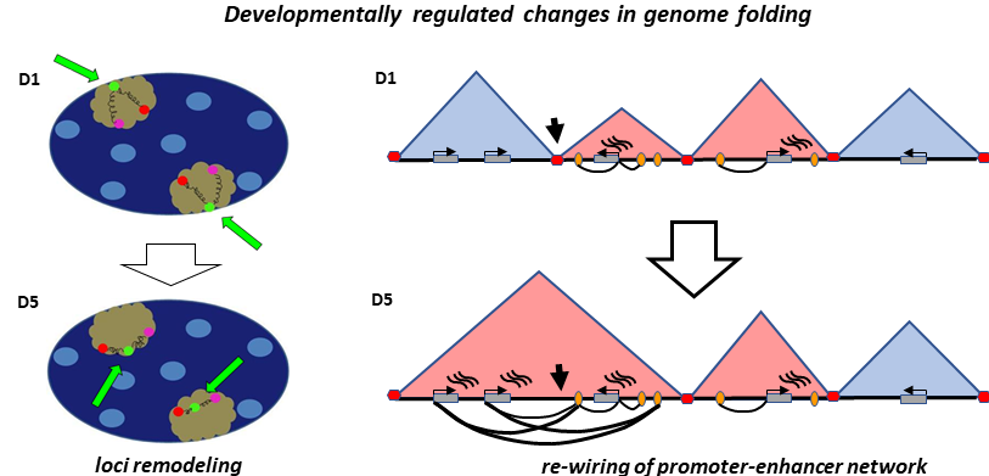
Our lab studies the role and mechanisms of epigenetic regulation in development, cancerogenesis and DNA replication. We are particularly interested how the 3D organization of the genome within the nucleus affects cell-type specific gene activity and DNA replication.
A typical mammalian genome consists of ~2-meter-long DNA that is contained in a nucleus with a diameter 200 000 times smaller. For proper cell functioning efficient, well organized 3D genome folding is essential. We study how distinct configurations of 3D nuclear architecture and chromatin folding regulate expression of distinct genes, re-wire promoter-enhancer networks. Furthermore, how they are integrated within developmental programmes.
Developmental Epigenetics Lab conducts inter-disciplinary research using a broad range of methods: classical molecular biology, cell engineering and animal models, combined with bioinformatics and advanced microscopy.
Selected publications
- The non-canonical SMC protein SmcHD1 antagonizes TAD formation and compartmentalization on the inactive X chromosome. Gdula MR*, Nesterova TB*, Pintacuda G, Godwin J, Zhan Y, Ozadam H, McClellan M, Moralli D, Krueger F, Green CM, Reik W, Kriaucionis S, Heard E, Dekker J, Brockdorff N, Nat. Comm. 2019, https://doi.org/10.1038%2Fs41467-018-07907-2
- Selective Roles of Vertebrate PCF11 in Premature and Full-Length Transcript Termination. Kamieniarz-Gdula K*, Gdula MR*(equal contribution), Panser K, Nojima T, Monks J, Wiśniewski JR, Riepsaame J, Brockdorff N, Pauli A, Proudfoot NJ. Mol Cell. 2019, https://doi.org/10.1016/j.molcel.2019.01.027
- PCGF3/5-PRC1 Initiates Polycomb Recruitment in X Chromosome Inactivation. Mafalda A, Pintacuda G, Masui O, Koseki Y, Gdula MR, Cerase A, Brown D, Mould A, Innocent C, Nakayama M, Schermelleh L, Nesterova TB, Koseki H, Brockdorff N, Science 2017 https://doi.org/10.1126/science.aal2512
- Pcf11 Orchestrates Transcription Termination Pathways in Yeast. Grzechnik P, Gdula MR, and Proudfoot NJ, Genes Dev. 2015 http://www.genesdev.org/cgi/doi/10.1101/gad.251470.114
- Spatial Separation of Xist RNA and Polycomb Proteins Revealed by Superresolution Microscopy, Cerase A, Smeets D, Tang YA, Gdula MR, Kraus F, Spivakov M, Moindrot B, Leleu M, Tattermusch A, Demmerle J, Nesterova TB, Green CM, Otte A, Schermelleh L, Brockdorff N, PNAS 2014 https://doi.org/10.1073/pnas.1312951111
- 5C Analysis of the Epidermal Differentiation Complex Locus Reveals Distinct Chromatin Interaction Networks between Gene-Rich and Gene-Poor TADs in Skin Epithelial Cells, Poterlowicz K, Yarker JL, Malashchuk I, Lajoie BR, Mardaryev AN, Gdula MR, Sharov AA, Kohwi-Shigematsu T, Botchkarev VA, Fessing MY, PLoS Genet. 2017 https://doi.org/10.1371/journal.pgen.1006966
- P63 and Brg1 Control Developmentally Regulated Higher-Order Chromatin Remodelling at the Epidermal Differentiation Complex Locus in Epidermal Progenitor Cells, Mardaryev AN, Gdula MR, Yarker JL, Emelianov VU, Poterlowicz K, Sharov AA, Sharova TY, Scarpa JA, Chambon P, Botchkarev VA and Fessing MY, Development 2014 https://doi.org/10.1242/dev.103200
- Remodeling of Three-Dimensional Organization of the Nucleus during Terminal Keratinocyte Differentiation in the Epidermis. Gdula MR, Poterlowicz K, Mardaryev AN, Sharov AA, Peng Y, Fessing MY, Botchkarev VA, J. Invest. Dermatol. 2013 https://doi.org/10.1038/jid.2013.66
- P63 Regulates Satb1 to Control Tissue-Specific Chromatin Remodeling during Development of the Epidermis. Fessing MY, Mardaryev AN, Gdula MR, Sharov AA, Sharova TY, Rapisarda V, Gordon KB, Smorodchenko AD, Poterlowicz K, Ferone G, Kohwi Y, Missero C, Kohwi-Shigematsu T, Botchkarev VA, J Cell Biol 2011 https://doi.org/10.1083/jcb.201101148
- Epigenetic Regulation of Gene Expression in Keratinocytes, Botchkarev VA, Gdula MR, Mardaryev AN, Sharov AA, Fessing MY, J. Invest. Dermatol. 2012 https://doi.org/10.1038/jid.2012.182
More publications in PubMed.