Home > Structure > Department of Gene Expression > Biomolecular Interactions and Transport
Biomolecular Interactions and Transport
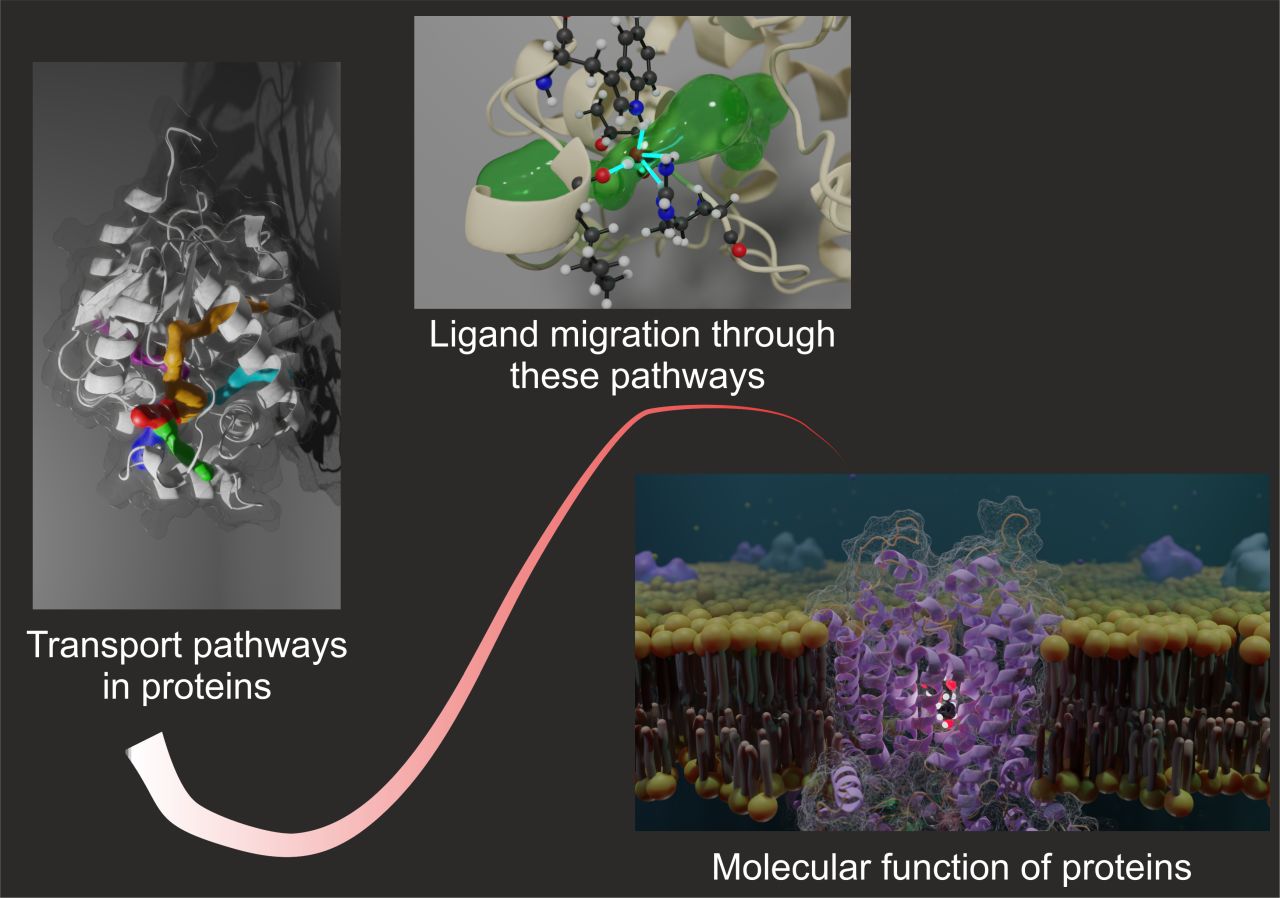
Our laboratory focuses on resolving the molecular details of ligand transport processes in proteins mainly relying on structural bioinformatics and computational simulations.

Prof. AMU Jan Brezovsky
Principal Investigator
Research
The main research topic of the laboratory is focused on solving fundamental questions about mechanisms involved in a ligand transport through proteins, and implications of such processes for the functions of living cells. To do so, we primarily employ various flavours of molecular dynamics simulations. To obtain a more comprehensive picture, we also investigate the protein-ligands interactions at the functional sites as well as intra- and inter-protein interactions. To achieve these goals, we also develop new computational protocols and tools and apply them to the analysis and design of biomedical and biotechnologically relevant proteins.
Selected publications
- Brezovsky J, Thirunavukarasu AS, Surpeta B, Sequeiros-Borja CE, Mandal N, Sarkar DK, Dongmo Foumthuim CJ, Agrawal N, 2022: TransportTools: a library for high-throughput analyses of internal voids in biomolecules and ligand transport through them. Bioinformatics 38: 1752-1753.
- Surpeta B, Grulich M, Palyzová A, Marešová H, Brezovsky J, 2022: Common Dynamic Determinants Govern Quorum Quenching Activity in N-Terminal Serine Hydrolases. ACS Catalysis 12: 6359-6374.
- Sequeiros-Borja CE, Surpeta B, Marchlewski I, Brezovsky J, 2022: Divide-and-conquer approach to study protein tunnels in long molecular dynamics simulations, MethodsX, in press
- Sequeiros-Borja CE, Surpeta B, Brezovsky J, 2021: Recent advances in user-friendly computational tools to engineer protein function. Briefings in Bioinformatics 22: bbaa150.
- Brezovsky J, Babkova P, Degtjarik O, Fortova A, Gora A, Iermak I, Rezacova P, Dvorak P, Kuta Smatanova I, Prokop Z, Chaloupkova R, Damborsky J, 2016: Engineering a De Novo Transport Tunnel. ACS Catalysis 6: 7597-7610.
- Bendl J, Musil M, Stourac J, Zendulka J, Damborsky J, Brezovsky J, 2016: PredictSNP2: A Unified Platform for Accurately Evaluating SNP Effects by Exploiting the Different Characteristics of Variants in Distinct Genomic Regions. PLoS Computational Biology 12: e1004962.
- Bendl J, Stourac J, Sebestova E, Vavra O, Musil M, Brezovsky J,* Damborsky J,* 2016: HotSpot Wizard 2: Automated Design of Site-Specific Mutations and Smart Libraries in Protein Engineering. Nucleic Acids Research 44: W479-487.
- Amaro M, Brezovsky J, Kovacova S, Sykora J, Bednar D, Nemec V, Liskova V, Kurumbang NP, Beerens K, Chaloupkova R, Paruch K, Hof M, Damborsky J, 2015: Site-Specific Analysis of Protein Hydration Based on Unnatural Amino Acid Fluorescence. Journal of the American Chemical Society 137: 4988-4992.
- Daniel L, Buryska T, Prokop Z, Damborsky J, Brezovsky J, 2015: Mechanism-Based Discovery of Novel Substrates of Haloalkane Dehalogenases using in Silico Screening. Journal of Chemical Information and Modeling 55: 54-62.
- Sykora J,* Brezovsky J,* Koudelakova T,* Lahoda M, Fortova A, Chernovets T, Chaloupkova R, Stepankova V, Prokop Z, Kuta Smatanova I, Hof M, Damborsky J, 2014: Dynamics and Hydration Explain Failed Functional Transformation in Dehalogenase Design. Nature Chemical Biology 10: 428-430.
More publications in PubMed.