Home > Structure > Human Molecular Genetics Research Unit
Human Molecular Genetics Research Unit
&
Laboratory of High Throughput Technologies
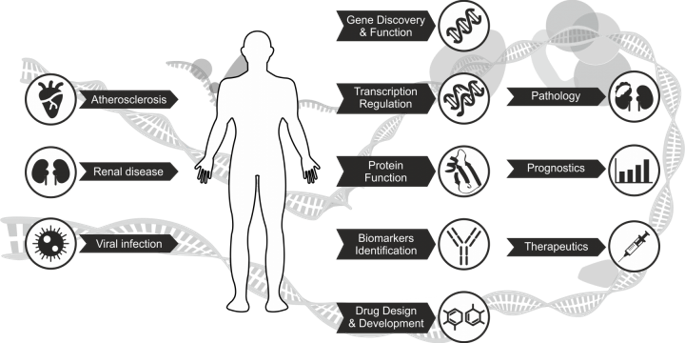
Our research concentrates on applications of integrative genomics and in silico modelling and virtual screening to understand, diagnose, and treat kidney cancer, atherosclerosis and infectious diseases

Prof. Hans Bluyssen
Principal Investigator

Prof. Joanna Wesoły
Principal Investigator
Research
LHMG aims at genome-wide and mechanistic understanding of the STAT-, IRF- and NF-kB-dependent signal integration between TLR4 and IFNs (Type I and II) in vascular and immune cells in experimental and clinical atherosclerosis. Group develops comparative virtual screening and docking validation tools to identify small inhibitory compounds of STATs and IRFs as a novel therapeutic strategy in the management of cardiovascular disease. Finally, LHMG aims at genome-wide understanding of canonical and alternative type I and II IFN-induced responses, to elucidate the timely Type I and II IFN-induced transcriptome and chromatin interactions mediated by STATs and IRFs, and identify known and novel protein coding ISGs and their potential antiviral functions.
LHTT aims at genomic characterization of clear cell Renal Cell Carcinoma (ccRCC), with the focus on transmembrane proteins (TMEMs) and their role in ccRCC etiology. Since molecular markers aiding early diagnosis and monitoring of ccRCC recurrence are virtually non-existent, LHTT concentrates on the identification of mutational and/or expression profiles, that can potentially mirror the stage of the disease in different body fluids.
Selected publications
- Time‑dependent recruitment of GAF, ISGF3 and IRF1 complexes shapes IFNα and IFNγ‑activated transcriptional responses and explains mechanistic and functional overlap Sekrecka A., Kluzek K., Sekrecki M., Eskandarian Boroujeni M., Hassani S., Yamauchi S., Sada K., Wesoly J., Bluyssen H. A. R. Cellular and Molecular Life Sciences 2023 https://doi.org/10.1007/s00018-023-04830-8
- Dysregulated Interferon Response and Immune Hyperactivation in Severe COVID-19: Targeting STATs as a Novel Therapeutic Strategy Eskandarian Boroujeni M., Sekrecka A., Antonczyk A., Hassani S., Sekrecki M, Nowicka H., Lopacinska N., Olya A., Kluzek K., Wesoly J., Bluyssen H. A. R. Frontiers in Immunology 2022, https://doi.org/10.3389/fimmu.2022.888897
- SINBAD: Structural, experimental and clinical characterization of STAT inhibitors and their potential applications Plens-Gałąska M., Woźniak T., Wesoły J. and Bluyssen H. A. R. Scientific data 2022, https://doi.org/10.1038/s41597-022-01243-3
- COVID-19 causes neuronal degeneration and reduces neurogenesis in human hippocampus. Bayat A.H., Azimi H., Hassani Moghaddam M., Ebrahimi V., Fathi M., Vakili K., Mahmoudiasl G.R., Forouzesh M., Eskandarian Boroujeni M., Nariman Z. and Abbaszadeh H.A.,. Apoptosis 2022 https://doi.org/10.1007/s10495-022-01754-9
- Identification of RCC Subtype-Specific microRNAs–Meta-Analysis of High-Throughput RCC Tumor microRNA Expression Data Kajdasz A., Majer W, Kluzek K., Sobkowiak J., Milecki T, Derebecka N, Kwias Z., Bluyssen H. A. R., Wesoly J. Cancers 2021, https://doi.org/10.3390/cancers13030548
- Signal Integration of IFN-I and IFN-II With TLR4 Involves Sequential Recruitment of STAT1-Complexes and NFκB to Enhance Pro-inflammatory Transcription. Piaszyk-Borychowska A., Széles L., Csermely A., Chiang H. C., Wesoły J., Lee C. K., Nagy L. & Bluyssen H. A. R. . Frontiers in immunology 2019,. https://doi.org/10.3389/fimmu.2019.01253
- Direct inhibition of IRF-dependent transcriptional regulatory mechanisms associated with disease Antonczyk A., Krist B., Sajek M., Michalska A., Piaszyk-Borychowska A., Plens-Galaska M., Wesoly J. & Bluyssen H. A. R. Frontiers in Immunology – Molecular Innate Immunity 2019, https://doi.org/10.3389/fimmu.2019.01176. eCollection 2019
- Thyroid cancers of follicular origin in genomic light – in depth overview of recently reported molecular marker candidates Pstrąg N., Ziemnicka K., Bluyssen H. A. R., Wesoły J. Molecular Cancer. 2018, https://doi.org/10.1186/s12943-018-0866-1
- Genetic characterization of polish ccRCC patients: somatic mutation analysis of PBRM1, BAP1 and KDMC5, genomic SNP array analysis in tumor biopsy and preliminary results of chromosome aberrations analysis in plasma cell free DNA Kluzek K., Srebniak M.I., Majer W., Ida A., Milecki T., Huminska K., van der Helm R.M., Silesian A., Wrzesinski T.M., Wojciechowicz J., Beverloo B.H., Kwias Z., Bluyssen H.A., Wesoly J. Oncotarget 2017, https://doi.org/10.18632/oncotarget.15331
- Identification of STAT1 and STAT3 specific inhibitors using comparative virtual screening and docking validation Szelag M., Czerwoniec A., Wesoly J., Bluyssen H.A. PLoS One 2015, https://doi.org/10.1371/journal.pone.0116688
More publications in PubMed.
Research grants
- Research Grant nr UMO-2012/07/B/NZ1/02710, 2013-2016: “STAT1 and IRF8-mediated signal integration between IFNg and TLRs: a pathophysiological role in vascular disease”. Project leader: Prof H. Bluijssen. National Science Center.
- Research Grant nr Preludium 2012/07/N/NZ2/01359, 2013-2016: „ Teoretyczny model SH2-zależnej dimeryzacji białka STAT1 jako nowe narzędzie do identyfikacji specyficznych inhibitorów aktywności”. Project leader: Mgr. Malgorzata Szeląg. Supervisor: Prof H. Bluijssen. National Science Center.
- Research Grant nr UMO-2013/11/B/NZ2/02569, 2014-2017: “Genome-wide characterization of type I IFN-induced transcriptomes and identification of novel ISGs and their antiviral functions”. Project leader: Prof H. Bluijssen. National Science Center.
- Research grant nr 2014/15/B/NZ2/00589 Mechanistic insight in Transmembrane Protein (TMEM) deregulation and its contribution to ccRCC etiology. 2015-2018 Project leader: prof. Joanna Wesoły. National Science Center.
- Research grant nr UMO-2014/13/B/NZ9/02123 Exploring genetic bases of innate and adaptive immune responses in chickens with next generation targeted DNA sequencing.
2015-2018. Participant: prof. Joanna Wesoły. National Science Center. - Research grant Odkryte na nowo. Kompleksowe opracowanie materiałów archeologicznych z neolitycznego stanowiska Kopydłowo 6, gm. Wilczyn.
02.2014-31.12.2015 Participant: Joanna Wesoły. National Heritage Board of Poland. - Research Grant nr UMO-2015/11/B/NZ2/02569, 2016-2019: Celowana inhibicja białek STAT i IRF jako nowa strategia terapeutyczna w leczeniu chorób układu sercowo-naczyniowego. Project leader: Prof H. Bluijssen. National Science Center.
- Research Grant nr UMO-2016/23/B/NZ2/00623, 2017-2021: Characterization of a novel intracellular amplifier circuit that controls long-term responsiveness to type I and type II IFNs. Project leader: Prof H. Bluijssen. National Science Center.
- Research Grant nr „PRELUDIUM” 2016/21/N/NZ2/01720, 2016/2019. The cooperation/role of unphosphorylated and phosphorylated ISGF3 components in the regulation of ISG expression and viral protection, Project leader: Mgr. Hanna Nowicka. Supervisor: Prof H. Bluijssen. National Science Center.
- Research grant nr 7/2020, 2020, Adam Mickiewicz University. Genome-wide characterization of immune responses in olfactory epithelial cells of COVID-19 patients as a measure of clinical outcome and treatment strategy. Project Leader: Prof. Hans Bluijssen. UAM.
- Research grant nr UMO-2020/37/B/NZ6/01080. NCN. The novel role of STAT1 in Vascular Smooth Muscle Cell and Macrophage common and –specific transcriptional responses that reflect onset and progression of atherosclerosis. Project Leader: prof. Hans Bluijssen. National Science Center.
Current group members

Prof. Hans Bluyssen
principal investigator
more…

Prof. Joanna Wesoły
principal investigator
more…

Katarzyna Kluzek, PhD
assistant professor
more…

Mahdi Eskandarian Boroujeni, PhD
postdoc
more…